 Jingdan Liang
Jingdan Liang
Associate professor
DNA phosphorothioation
Email: jdliang@sjtu.edu.cn
DNA Phosphorothioate Modification as an Antioxidant
Endogenous superoxide and hydrogen peroxide are generated as by-products of aerobic metabolism. Reactive oxygen species scavenging enzymes and small molecules have been being evolved to protect cells. A newly finding is, diverse bacteria contain DNA with sulfur incorporated stereo-specifically into their DNA backbone, called phosphorothioate modification. Phosphorothioate DNA can react with hydrogen.
The phosphorothioate DNA hosting bacteria show increasing resistance to hydrogen peroxide. Inside the cells, phosphorothioate DNA is more resistant to the double-strand break damage caused by hydrogen peroxide than phosphorothioate-free DNA. These findings are consistent with a hypothesis that phosphorothioation modification endows DNA with reducing chemical property, protecting the bacteria against reactive oxygen species.
The molecular damages accumulated from reactive oxygen species is one of the causes leading to aging and diseases in human. The newly finding that phosphorothioate modification function as an antioxidant provides a novel tool to probe the cause and effect relationship of DNA oxidative damage and the related human disease.
Research Interests
1. Bacterial DNA phosphorothioate modification as an antioxidant
2. The application of DNA phosphorothioation as a tool
DNA modification, most often in the form of methylation, occur naturally both in prokaryotes and in eukaryotes. Subtle changes at bases, affected by proteins recognizing modified/unmodified form of specific nucleotide sequences, have profound impact on a variety of physiological aspects of related organisms. In bacteria, DNA methylation can serve as a restriction and modification system to defend the host from phage, as a genetic switch to turn specific gene expression on or off, and as a marker to coordinate chromosome replication and mismatch repair.
Although DNA backbone phosphodiester linkage is structurally monotonous, potential alterations, as a playground of chemists, have been extensively explored. DNA phosphorothioation, in which the non-bridging oxygen swapped with sulfur, is one of such effective alteration. Owing to its property of resistance to nuclease digestion, DNA phosphorothioation is widely used in biochemical and clinical applications. Interestingly, bacteria also have the ability to harvest the potential of such DNA alteration: the once proved to be sulfur containing DNA modification turns out to be DNA phosphorothioation. Physiological DNA phosphorothioation raised many questions, as noted by Professor Fritz Eckstein: 1, how and why has nature chosen this particular modification? 2, what could be the mechanism of post-replicative change of phosphate to phosphorothioate? 3, how sites of DNA thiolation are chosen? 4, what are the function and advantages of this phosphorothioate modification? Currently, I'm concentrating on related questions.
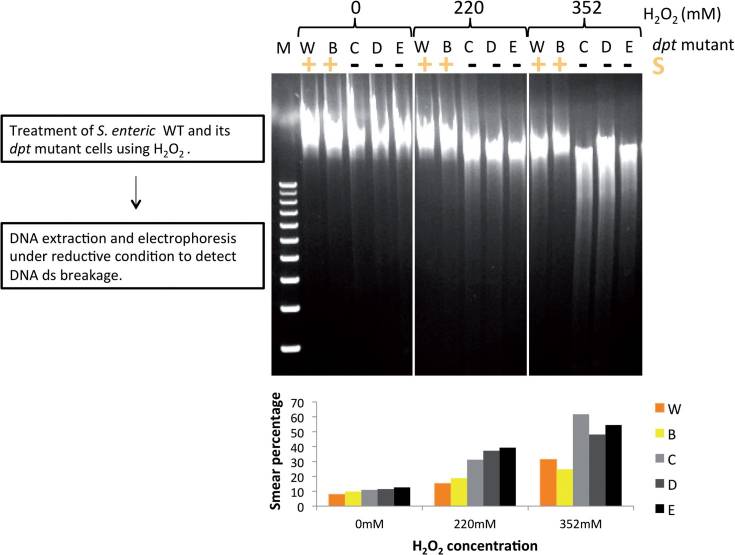
Figure 1 In vivo resistance of PT DNA to double-strand breakage caused by H2O2 Salmonella cells were incubated with increasing concentration of H2O2 for 2h. (Xie, et al. Nucleic Acids Res. 2012 )
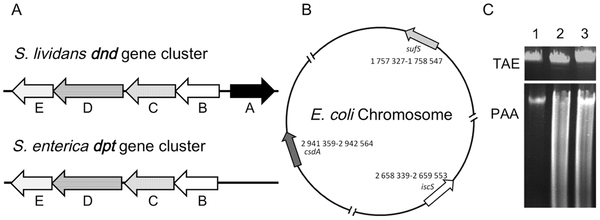
Figure 2 Heterologous expression of the S. enterica serovar cerro 87 dptBCDEgenes in E. coli BW25113. (An, et al. PLoS One. 2012)
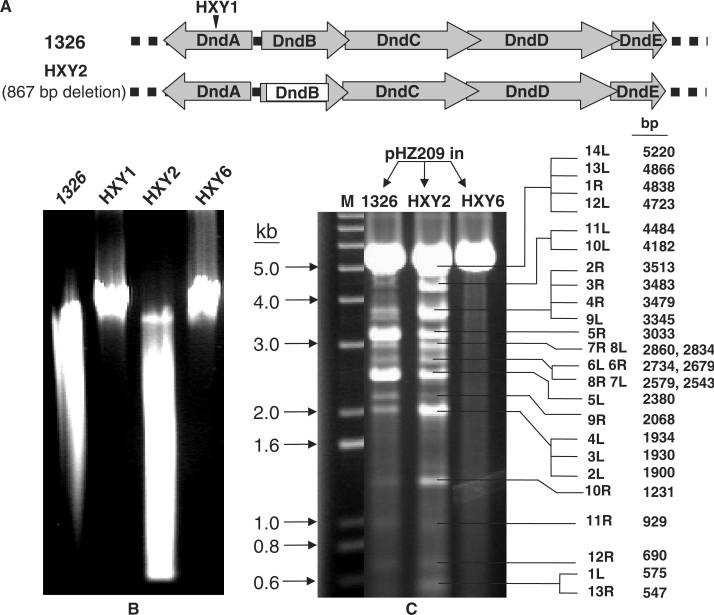
Figure 3 (A) Organization of the dnd gene cluster and in-frame deletion of dndB. (B) Enhanced Dnd phenotype displayed by dndB− mutant HXY2 as compared with that of wild-type 1326. Lack of Dnd phenotype displayed by dndA− mutant HXY1 and dnd-deletion mutant HXY6 is shown as controls. (C) Preferential cleavage at specific site of EcoRV-linearized pHZ209 originating from 1326, but more random and lack of cleavage of the same DNA originating from HXY2 and HXY6, respectively. All the samples were treated with activated Tris-buffer followed by gel electrophoresis. (Liang, et al. Nucleic Acids Res. 2007)
Education Experience
2011-2013 Visiting scholar,
The Department of Molecular & Cell Biology (MCB), University of California, Berkeley
2003-2007 Ph.D. in Microbiology, School of Life Sciences and Biotechnology, Shanghai Jiaotong University,
With Prof. Zixin Deng.
1994-1997 M.S. in Molecular Plant Pathology, Department of Plant Pathology, Nanjing Agricultural University,
With Prof. Jinsheng Wang.
1990-1994 B.S. in Microbiology, University of Shanxi.
Work Experience
2012- Tutor for graduate, SJTU, China
2011- Associate Professor, Tutor for graduate, SJTU, China
2008- Assistant professor, Key Laboratory of Microbial Metabolism,Ministry of Education,
School of Life Sciences and Biotechnology, Shanghai Jiaotong University.
1997-2003 Lecturer Department of Chemical Engineering,Nantong University
Selected Publications
1. Phosphorothioate DNA as an antioxidant in bacteria. Xie XQ*, Liang JD*, Pu TN, Xu F, Yao F, Yang Y, Zhao YL, You DL, Zhou XF, Deng ZX, Wang ZJ. Nucleic Acids Research. 2012 Oct; 40(18):9115-24. (*These authors contributed equally to this work)
2. A novel target of IscS in Escherichia coli: participating in DNA phosphorothioation. An XH, Xiong W, Yang Y, Li FH, Zhou XF, Wang ZJ, Deng ZX*, Liang JD*. PLoS One. 2012; 7(12):e51265. (*Corresponding author).
3. Effective elimination of nucleic acids from bacterial protein samples for optimized blue native polyacrylamide gel electrophoresis. Liang JD*, Niu QL*, Xu XP, Luo YM, Zhou XF, Deng ZX, Wang ZJ. Electrophoresis. 2009 Jul; 30(14):2454-9. (*These authors contributed equally to this work)
4. DNA phosphorothioation in Streptomyces lividans: Mutational analysis of the dnd locus. Xu TG*, Liang JD*, Chen S, Wang LR, He XY, You DL, Wang ZJ, Li AY, Xu ZL, Zhou XF, Deng ZX. BMC Microbiol. 2009 Feb 20; 9(1):41. (*These authors contributed equally to this work)
5. DNA modification by sulfur: analysis of the sequence recognition specificity surrounding the modification sites. Liang JD*, Wang ZJ*, He XY, Li JL, Zhou XF,Deng ZX. Nucleic Acids Research. 2007 Apr; 35(9):2944-54. (*These authors contributed equally to this work)
6. Characterization of the N-methyltransferase CalM involved in calcimycin biosynthesis by Streptomyces chartreusis NRRL 3882. Wu Q, Gou L, Lin S, Liang J, Yin J, Zhou X, Bai L, An D, Deng Z, Wang Z. Biochimie. 2013 Apr 11.
7. Mutasynthesis of Pyrrole Spiroketal Compound by Using Calcimycin 3-Hydroxy Anthranilic Acid Biosynthetic Mutant. Gou L, Wu Q, Lin S, Li X, Liang J, Zhou X, An D, Deng Z, Wang Z. Applied Microbiology and Biotechnology , 2013 May 12.
8. Characterization of the Pyrrole Polyether Antibiotic Calcimycin (A23187) Biosynthetic Gene Cluster from Streptomyces chartreusis NRRL 3882. Wu Q, Liang J, Lin S, Zhou X, Bai L , Deng Z and Wang Z. Antimicrobial Agents and Chemotherapy (AAC),2011 Mar; 55(3):974-82.
9. Theoretical study on steric effects of DNA phosphorothioation: B-helical destabilization in Rp-phosphorothioated DNA. Zhang Y, Liang J, Lian P, Han Y, Chen Y, Bai L, Wang Z, Liang J, Deng Z, Zhao YL. J Phys Chem B.2012 Sep 6;116(35):10639-48.
10. Crystal structure of the cysteine desulfurase DndA from Streptomyces lividans which is involved in DNA phosphorothioation. Chen F, Zhang Z, Lin K, Qian T, Zhang Y, You D, He X, Wang Z, Liang J, Deng Z, Wu G. PLoS One. 2012; 7(5):e36635.
11. Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification. Hu W, Wang C, Liang J, Zhang T, Hu Z, Wang Z, Lan W, Li F, Wu H, Ding J, Wu G, Deng Z, Cao C. Cell Res.2012 Jul;22(7):1203-6.
12. Purification, crystallization and preliminary X-ray analysis of the DndE protein from Salmonella enterica serovar Cerro 87, which is involved in DNA phosphorothioation. F. Chen, K. Lin, Z. Zhang, L. Chen, X. Shi, C. Cao, Z. Wang, J. Liang, Z. Deng, G. Wu. Acta Crystallogr Sect F Struct Biol Cryst Commun.2011 Nov 1; 67(Pt 11):1440-2.
13. Two pHZ1358-derivative Vectors for Efficient Gene Knockout in Streptomyces. He, Y, Wang Z, Bai L, Liang J, Zhou X, Deng Z. J. Microbiol. Biotechnol. 2010 4; 20(4): 678-682
Copyright © 2016,The Laboratory of Molecular Microbiology at Shanghai Jiao Tong University. All rights reserved.
Add:Science Building, Xuhui Campus, Shanghai Jiaotong University, Shanghai 200030, China. Tel:021-62932943
